

Medicinal chemist’s insights into the microbiome
The human microbiota consists of trillions of microorganisms, including bacteria, yeasts, and viruses, which coexist in various parts of the body including the skin, lungs, and most notably, the gut. In particular, the gut microbiome has attracted significant attention in recent years due to its impact on human health and disease.1 This unique community of microorganisms could have a major impact on the efficacy and toxicity of drugs, particularly those administered orally. This influence stems from the ability of gut microbes to metabolize drugs, potentially leading to the formation of toxic metabolites and affecting absorption, distribution, and elimination of pharmacologically active compounds.2
Just as hepatic stability is assessed for drug development, the microbiota should be given the same attention.
In this context, it’s becoming essential to include the microbiome as a key player in drug development, much like the liver is traditionally considered in drug metabolism. While we have well-defined protocols for assessing hepatic stability across different species (i.e in vitro hepatic clearance from mouse, rat, dog, monkey or human hepatocytes; IVIVE) the variability of the microbiome is a challenge for drug development. In fact, microbiota composition is not static within an individual but can change based on factors like environment, period of the year, life stage, hormones, and diet.3
In drug development, the variations in microbiota could potentially contribute to the differing drug responses observed in some patients (Scheme I). One patient’s microbiota may contain bacteria that metabolize the drug into an inactive form, reducing treatment efficacy. In contrast, another patient’s microbiota will produce another metabolite leading to adverse effects.
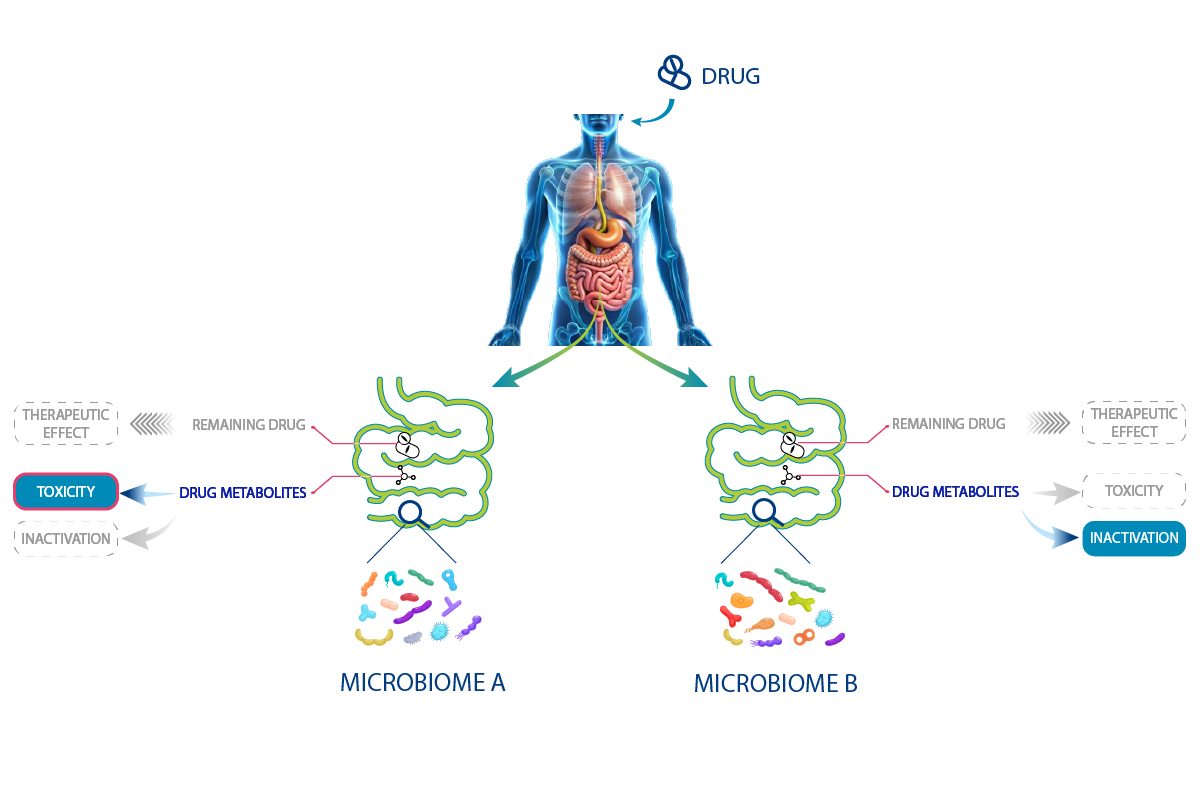
Scheme I – As many treatment responses as different microbiota
Some of these interactions between drugs and microbes are already known, for exemple the interaction between Levodopa (L-DOPA) and Enterococcus faecalis bacteria for the treatment of Parkinson’s disease.4 Indeed, these bacteria produce tyrosine decarboxylase enzymes that convert L-DOPA into dopamine, preventing it from crossing the blood-brain barrier and making supplementation ineffective. In addition, the dopamine may be converted into m-tyramine by another gut bacteria, Eggerthella lenta, which could lead to adverse effects.5 This cascade of enzymatic reactions can be reduced by combining Levodopa administration with an inhibitor of tyrosine decarboxylase.6
For precision medicine, analyzing the microbiota in individual patients could allow for tailored treatments by co-administering the drug with probiotics or other beneficial microorganisms. This insight is relevant not only to approved drugs but also, and most importantly, to the development of new drug candidates from early stages. By considering the microbiome at the very beginning of drug development process, medicinal chemists could identify potential side effects resulting from microbiome-mediated drug metabolism before they become an issue.
Strategies to include the gut microbiota in drug development
Today, the primary limitation to integrating the microbiome into early drug development is the limited availability of data on drug-microbiome interactions, although it is constantly improving. A recent review written by Medicen (a French human health innovation cluster combining experts from pharmaceutical companies, academia, biotechs, clinical research organizations, and regulatory science) provides an overview of available microbiome-related tools.7
As illustrated in Scheme II, it could be worthwhile to conduct parallel studies of microbiome-drug interactions and traditional metabolic stability assays. At the lead optimization phase, in silico databases8 and in vitro screening assays9 are being developed to investigate the effect of specific gut microbiota on the metabolism of drugs. In vivo models, including germ-free rodents (without microbiota) or germ-free rodents colonized with specific bacteria are emerging as valuable tools to assist medicinal chemists in more advanced stages of the drug design process.
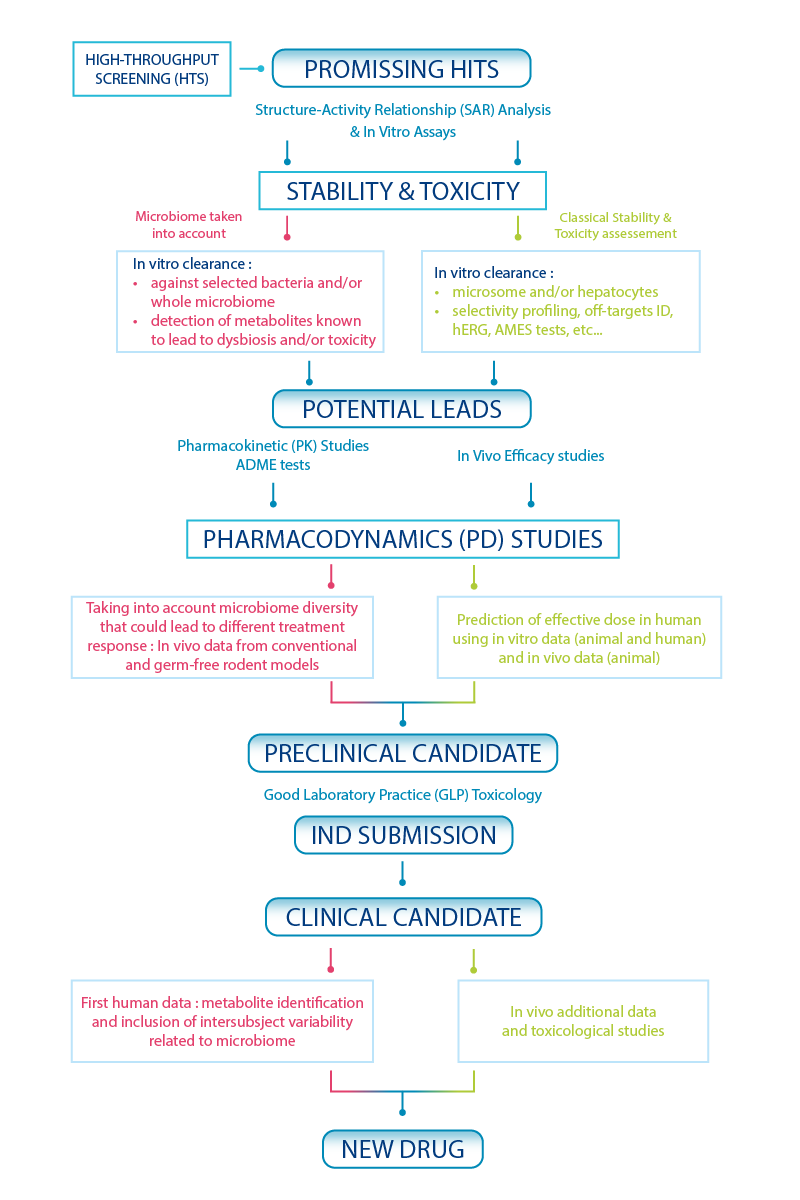
Scheme II – Drug discovery screening cascade including microbiome-based assays
Seeking support from an experienced team for your R&D programs?
Building on our expertise at Oncodesign Services, we offer customized solutions for studying the efficacy of bacteria-based therapies, as well as the impact of drugs on the microbiota using both in vitro and/or in vivo studies. These models were successfully used for in vivo evaluations of bacteria for immuno-oncology research10.
Tailoring treatments based on individual microbial profiles holds promise for improving therapeutic outcomes and reducing adverse effects. In drug development, the variations in microbiota could potentially contribute to the differing drug responses seen in some patients’ population. New methods are being developed to incorporate the microbiome. These models, including in silico microbiome–drug interaction databases but also in vitro and in vivo studies, position the microbiome as a metabolic network similarly to the liver. By studying the microbiome, medicinal chemists could anticipate drug side effects early in development and before entering clinical phase.
About the author
This article was written by Audrey Dumoulin, medicinal chemist at Oncodesign Services. In her current role, Audrey is involved in supporting integrated projects for customers.
A question? A project around Microbiome? Contact us!
-
References
[1] Koppel, N. et al. Science 2017, 356, 6344, eaag2770. Chemical transformation of xenobiotics by the human gut microbiota. DOI:10.1126/science.aag2770.
[2] Zhao, Q. et al. Sig. Transduct. Target Ther. 2023, 8, art. n°386. Drug-microbiota interactions: an emerging priority for precision medicine. DOI:10.1038/s41392-023-01619-w.
[3] Zimmermann, M. et al. Nature 2019, 570, 462. Mapping human microbiome drug metabolism by gut bacteria and their genes. DOI:10.1038/s41586-019-1291-3.
[4] Van Kessel, S.P. et al. Nat Commun 2019, 10, 310. Gut bacterial tyrosine decarboxylases restrict levels of levodopa in the treatment of Parkinson’s disease. DOI:10.1038/s41467-019-08294-y
[5] Rekdal, V. M. et al. Science 2019, 364, eaau6323. Discovery and inhibition of an interspecies gut bacterial pathway for Levodopa metabolism. DOI:10.1126/science.aau6323.
[6] Porter, C. C. et al. Biochemical Pharmaco. 1962, 11, 1067. Inhibition of dopa decarboxylase by the hydrazino analog of α-methyldopa. DOI: 10.1016/0006-2952(62)90166-1.
[7] Medicen Microbiome Drug Metabolism Working Group. Drug Metab Dispos. 2024, 52, 4, 274. Gut Microbiome Integration in Drug Discovery and Development of Small Molecules. DOI: 10.1124/dmd.123.001605.
[8] Yin, J. et al. Nucleic Acids Res. 2021, 8, 49, 1233. INTEDE: interactome of drug-metabolizing enzymes. DOI: 10.1093/nar/gkaa755.
[9] Van Leeuwen, P. T. et al. FEMS Microbiol. Rev. 2023, 47, 2, fuad012. Synthetic microbial communities (SynComs) of the human gut: design, assembly, and applications. DOI: 10.1093/femsre/fuad012.